All published articles of this journal are available on ScienceDirect.
Production of Cellulase Enzymes by Phlebiopsis sp. Cultured on Untreated and Pretreated Sugarcane Bagasse, Maize Cobs, and Rice Husks
Abstract
Background/Introduction
Clean energy from renewable and dependable sources is in great demand around the world. Cellulose is one of the most ubiquitous organic components often broken down by cellulase to produce biofuels. Fungi that reside in the environment can be used to produce cellulase enzymes.
Methodology
This study focused on the isolation, identification, and characterization of Phlebiopsis sp. from a decaying tree trunk and exploring untreated and pretreated maize cobs, sugarcane bagasse, and rice husks as substrates for cellulase production under solid state fermentation in sterilized bottles at 24°C room temperature in a dark place. The crude enzyme collected after fermentation was used to carry out total cellulase (FPase), exoglucanase, endoglucanase, and β-glucosidase activities.
Results
For the untreated substrates, Phlebiopsis sp. produced the highest endoglucanase when cultured on sugarcane bagasse at 74.3±0.27 IU/mL. Pretreatment of maize cobs with 0.25M NaOH resulted in the highest exoglucanase and endoglucanase activity by Phlebiopsis sp. at 6.17±0.04 IU/mL and 25.1±0.61 IU/mL, respectively. The overall results showed that untreated substrates produced a higher cellulase activity than pre-treated substrates except β-glucosidase activity. The yielded β-glucosidase activity by Phlebiopsis sp. was the highest when maize cobs were pretreated with hot water at 118 ± 0.42 IU/mL and 0.1m HCL at 118 ± 0.44 Ul/mL.
Discussion
Phlebiopsis sp. effectively produced cellulases from agro-waste, with untreated sugarcane bagasse and maize cobs yielding the highest activity. These findings support its potential for low-cost biofuel production and align with existing research on fungal cellulases. Limited resources, however, constrained deeper analysis and broader validation.
Conclusion
This study reveals that Phlebiopsis sp. is a cellulose-degrading fungus that can be used for cellulase enzyme production. Moreover, maize cobs and sugarcane bagasse are good carbon sources for cellulase enzyme production but further analysis should be done on the appropriate pretreatment methods to be used to increase cellulase production.
1. INTRODUCTION
Currently, the global community’s main areas of concern are the environment and energy. The increasing population across the world, accompanied by rapid industrial growth, has escalated the demand for energy [1]. The extreme use of fossil fuels has motivated scientists to figure out other alternative sources for renewable biofuel production [2]. A huge amount of agricultural waste is produced yearly, making it useful for the generation of cleaner biofuels at an affordable cost [3]. Lignocellulosic Biomass (LCB) consists of energy-rich biopolymers capable of contributing to global fuel demands in a renewable and sustainable manner [4]. LCB is made of cellulose (40-50%), hemicelluloses (15-20%), and lignin (20-30%) as constituents of the plant cell wall organized in a convoluted non-uniform three-dimensional structure [4]. The largest renewable biological compound on terrestrial earth is cellulose from the polysaccharide class of phytochemicals composed of linear β-1,4-linked D-glucopyranose chains, which are broken down by cellulases [5]. Fungi, bacteria, and protozoa are the microorganisms known to break down cellulose, hemicellulose, and lignin. Fungi degrade lignocellulose using a complex and effective mechanism, consisting of a collection of different enzymes [6]. The endo-β-1, 4-D-glucanase mediates the breakdown of internal glycosidic bonds of respective individual cellulose chains resulting in the exposure of cellulosic constituents of polysaccharide, which can then be processed by the exo-β-1, 4-D-glucanase into two to four separate glucose units giving rise to disaccharides including cellobioses. The final step of β-D glucosidase activity results in the formation of D-glucose units via the hydrolysis of cellobiose [7].
There has been an increase in studies in relation to fungi that produce cellulases, which could be used in the generation of chemical feedstock by biotransformation of plant and industrial wastes [8]. The cost of the cellulases needed for biomass conversion is high since they make up a significant portion of the overall process cost [9]. Therefore, there is a need to explore sources of cheap and effective cellulolytic enzymes critical for the production of biofuels, most importantly, those capable of converting lignocellulose into fermentable sugars [10].
The creation of straightforward pretreatment techniques that will successfully delignify a variety of lignocellulosic biomass substrates is essential for the success of cellulase enzyme manufacturing [11]. Since lignocellulosic materials differ in their physico-chemical properties, it is essential to apply pretreatment techniques appropriate for each raw material used for producing cellulases [12]. Methods for chemical pretreatment include oxidative delignification, acid or alkaline hydrolysis, and ozonolysis [13, 14]. Cellulose digestibility is increased by alkali pretreatments, which leads to lignin solubilization but alkali pretreatment exhibits minor cellulose and hemicellulose solubilization than acid pretreatment [15]. A suitable reagent for alkali pretreatment is sodium hydroxide. Sodium hydroxide's function is to break lignin by causing swelling, which increases cellulose's internal surface and decreases crystallinity and polymerization levels. Enzyme production will improve with the removal of lignin sites which render cellulose inaccessible by key enzymes involved in its hydrolysis. Initial exposure of alkali treatment has proven more effective on agricultural residues compared to woody components [16]. The main focus of this particular study was to isolate and identify Phlebiopsis sp. and explore its cellulase enzyme production by the utilization of the readily available lignocellulose substrates, such as maize cobs, sugarcane bagasse, and rice husks. Sustainable biofuel production from lignocelluloses on an industrial scale therefore, directly depends on the ability to produce this valuable enzyme from the above, cheap and renewable raw sources. In this study, the effect of incubation time in the production of cellulosic enzymes was determined by the use of both untreated and pre-treated rice husks, maize cobs, and sugarcane by solid-state fermentation using Phlebiopsis sp.
2. METHODOLOGY
2.1. Collection of Samples
Wood samples from different tree trunks were collected from a forest located at coordinates 1°15'26.0”S 36°44'53.2” E in Nairobi, Kenya. The total number of samples collected was 20. The samples were ferried to the study site laboratory at Kenyatta University at the Biochemistry, Microbiology, and Biotechnology Department and stored under 25°C (normal room temperature) until further processing.
2.2. Preparation of 1% Carboxymethyl Cellulose (CMC) Agar
Carboxymethylcellulose agar was prepared according to Legodi et al., [17] by adding the following (g/L): 1.88 of carboxymethyl cellulose, 1.0 of Glucose, 15 of Agar powder, and 10ml of stock solution (1% Congo red). The final solution was made up by the addition of 1 liter of distilled water. The prepared agar medium was then subjected to autoclaving at 121°C and pressure of 15 psi for 20 minutes, after which 2.5 ml of gentamycin antibiotic (100 mg/L) was added before dispensing the media into sterile plastic Petri dishes [18].
2.3. Screening and Isolation of Cellulose-degrading Fungi
The decomposing wood particles were sectioned into 1cm pieces. For each sample, surface sterilization was carried out using a 1% sodium hypochlorite solution for 1 minute. The sterilized samples were then rinsed three times with sterile distilled water and blot-dried with sterile paper towels. The samples were then subsequently plated on 0.188% Carboxymethyl cellulose (CMC) agar media supplemented with gentamycin (100 mg/L) to prevent bacterial growth. The Petri dishes were labeled, sealed with parafilm, and incubated at 25°C (room temperature). Monitoring and recording of fungal growth on respective plates was done daily. Emerging colonies were sub-cultured into CMC-Congo red media for isolation of cellulose-degrading fungi. The isolated fungi that showed zones of clearance on CMC agar were cellulase-producing fungi. The zone of clearance was due to the hydrolysis of cellulose present in the agar plate by the cellulase enzyme released by the fungi.
2.4. Preparation of Potato-dextrose Agar (PDA)
For potato dextrose agar preparation, the protocol by Westphal et al. [19] was used. Irish potatoes weighing 200 g were chopped into small sizable pieces and introduced into a 1000 mL conical flask. The conical flask was filled with 500ml of distilled water and covered with aluminum foil. The solution was boiled on a hotplate. After boiling, the broth was decanted into another 1000 mL conical flask. Twenty (20) g of dextrose was added to the broth. Distilled water was added to reach 1000ml and 15 g of agar was added. The media was sterilized for 20 minutes at 121°C, 15 psi. Gentamycin was added when the media had cooled to 45°C to prevent bacterial growth before dispensing into sterile plastic petri dishes.
2.5. Subculturing Cellulolytic Fungi
Pure sample cultures with zones of clearance on the CMC agar plates were sub-cultured on other PDA plates. The plates were incubated in a dark place at room temperature for 1 week to monitor fungal growth.
2.6. Molecular Identification of Lignocellulolytic Fungi
2.6.1. Isolation of Genomic DNA
Extraction of the total genomic DNA was done from plates containing pure fungal cultures with zones of clearing on CMC agar, which were then sub-cultured and grown on PDA [20]. Sterile scalpels were used to extract fungal mycelium and liquid nitrogen was used to grind it to powder form with a mortar and pestle. Using a sterile spatula, about 300 mg of the powder was transferred to 2ml sterile microcentrifuge tubes. To each tube, 400 µl of the extraction buffer, which includes 3% CTAB, 100 mM Tris-HCl (pH 7.5), 1.4 M NaCl, 20 mM EDTA (pH 8.0), 2% polyvinylpyrrolidone, and 1% mercaptoethanol (pre-heated to 65°C) was added. The microfuge tubes' contents were well mixed before being incubated in a water bath at 65°C for 45 minutes. The tube was cooled down to 25°C for 5 minutes before adding 400 mL of chloroform/isoamyl alcohol (24:1 v/v) and let it to stand for 15 minutes at room temperature. Centrifugation of the mixture was done for 15 minutes at 13,200 RPM. The aqueous layer was transferred to newly labeled 1.5 ml microcentrifuge tubes. To precipitate the genomic DNA, 0.7 liters of ice-cold isopropanol were added to the supernatant. The contents of the tubes were mixed by inverting them twenty-five times before incubating them at -20°C for three hours or overnight. The contents of the tubes were centrifuged at 13,200 rpm. The pellets were rinsed with 300 µL of ice-cold 70% ethanol and centrifuged at 13,000 rpm for one minute. The surplus alcohol was removed, and the tubes were inverted inside a lamina flow to let the pellets air dry. The dried pellets were re-suspended in 50 µL of TE buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA (pH 7.5)], and 3 µL of RNase-A (10 mg/mL) was added. The tubes were incubated at 37°C for 30 minutes. The quality of the extracted genomic DNA was assessed using 0.8% agarose gel electrophoresis [21] and kept at -20°C for future use.
2.6.2. PCR Amplification
The 18S rRNA of internally transcribed fungal DNA sections was amplified using ITS4R (TCCTCCGCTTATTGATATGC) and ITS86F (GTGAATCATCGAATCTTTGAA) primers [22] in a PCR thermocycler. A 22 µL PCR reaction mixture was employed, which included 10.6 µL of sterile ddH2O, 2 µL of forward primer, 2 µL of reverse primer, 4 µL of MyTaq buffer, 0.4 µL of MyTaq polymerase (Bioline, USA), and 3 ng of template DNA. The PCR conditions were as follows: 1 cycle of 95°C initial denaturation for 3 minutes; 35 cycles of 95°C denaturation for 30 seconds, 55°C annealing for 30 seconds, and 72°C extension for 1 minute; 1 cycle of 72°C final extension for 10 minutes; and a 4-minute holding stage at 15°C.
2.6.3. PCR Products Resolution and Sequencing
The PCR products were separated with 1.2% agarose gel electrophoresis. The gel electrophoresis was carried out according to Babolhavaeji et al. [23]. 1.2 grams of agarose were mixed into 100 mL of TBE buffer and heated in the microwave for 30 seconds. 0.5 µg/mL of ethidium bromide was added before the melted agarose was poured into a casting tray. The gel was put in a chamber containing TBE buffer, with the wells near the negative electrode of the chamber. The gel was loaded with a 1kb DNA ladder dyed in sybr green as a size reference. Sybr green was used as the loading dye and the gel run for 45 minutes at 100V before the results were viewed under UV light. The PCR products were sent to Macrogen Inc. (Netherlands) for sequencing utilizing the Sanger dideoxy sequencing method.
2.7. Screening of Agro-wastes as Substrates for Cellulase Production
2.7.1. Experimental Design
A multifactorial experimental design was used to investigate the suitability of untreated and pretreated maize cobs, rice husks, and sugarcane bagasse for the production of cellulase enzymes using Phlebiopsis sp. To ensure repeatability, the tests were done in triplicates. The independent factors for Phlebiopsis sp. cellulase enzyme synthesis were the three substrates and four incubation time, whereas the dependent variable was cellulolytic enzymes (filter paper, endoglucanase, exoglucanase, and β-glucosidase).
2.7.2. Cellulosic Substrates
Sugarcane bagasse, maize cobs, and rice husks were screened for suitability for cellulase production under SSF. The cellulosic substrates were ground using an electric mill and sieved through a 2 mm sieve.
2.7.3. Fungal Inoculum
PDA liquid medium was made according to the technique of Hasanini et al. [24], without the agar, as indicated in 2.4 above. The medium was sterilized for 20 minutes at 121°C and 15 pressure. The medium was transferred to sterile 250 ml conical flasks. In each sterile flask, 50 mL of potato dextrose liquid medium was added. A sterile scalpel was utilized to transfer pure Phlebiopsis sp. cultures grown on PDA to the liquid medium. The liquid medium was cultured at 25°C room temperature on a shaker at 150 rpm [25] for one week while the fungi's growth was monitored.
2.7.4. Pretreatment of Cellulosic Substrates
For pre-treated substrates, 400 g of each of the substrates was pre-treated with distilled water, 0.25 M NaOH, 0.1 M H2SO4, and 0.1 M HCL at a temperature of 121°C and pressure 15 psi for 15 minutes thereafter; each substrate was washed in running water until the pH was 7.0.
2.7.5. Solid State Fermentation
Ten (10) ml of Bushnell Haas medium solution (g/l) [0.1 g of MgSO4.7H2O, 0.5 g of Na2HPO4, 0.5 g of KH2PO4, 0.5 g of NH4NO3, 0.05 g of FeCl3. 6H2O and 0.02 g of CaCl2] were used as a wetting agent and added to 350 mL bottles with 5g of sugarcane bagasse, 5g of rice husks, and 5 g of maize cobs before the bottles were autoclaved at a temperature of 121°C and pressure 15 psi for 15 minutes [26]. After cooling, substrates were inoculated with a 2 ml inoculum containing 1 x 107 spores/ml, which was prepared as in section 2.7.3 and incubated at room temperature. Samples were taken every 3 days for the untreated substrates to determine the effect of incubation time on cellulase enzyme production, while for pretreated substrates a single extraction was done after 12 days of incubation at room temperature.
2.7.6. Extraction of Crude Enzymes
In order to extract cellulolytic enzymes, 50 mL of 50 mM citrate buffer pH 4.8 was added to the bottles. The bottles were shaken and left for two hours to allow the enzymes to extract into the buffer solution. After cheesecloth filtration, the liquid extract from the bottles was centrifuged at 10,000 xg for 10 minutes at 25°C room temperature. The supernatant was collected and stored -20°C for biochemical tests of total cellulase (FPase), endoglucanase, exoglucanase, and β-glucosidase enzyme activities.
2.8. Determination of Cellulase Activity
2.8.1. Preparation of Dinitrosalicylic Acid (DNS) and Potassium Sodium Tartrate
Dinitrosalicylic acid (DNS) solution was made by dissolving 10 g of 3,5-Dinitrosalicylic acid, 10 g of sodium hydroxide, 2 g of phenol, and 0.5 g of sodium sulphite in 1000 mL of distilled water. To prevent light exposure, the solution was kept in a dark container for two weeks. Potassium sodium tartrate solution was made by dissolving 400 g of potassium sodium tartrate in 1000 mL distilled water.
2.8.2. Filter Paper Assay
The total cellulase activity was evaluated in triplicate using the filter paper assay (FPA) technique [27]. The Whatman No. 1 filter paper was cut into 1cm × 6 cm strips, submerged in 500 µL of 50 mM sodium citrate buffer pH 4.8 in a 2ml microcentrifuge tube, and 500 µL aliquots of crude enzyme extract added [28]. After an hour of incubation at 50°C, the reducing sugars produced were measured using the Dinitrosalicylic Acid (DNS) approach [29]. To halt the reaction, add 700 µL of DNS reagent, then immerse the tubes in boiling water for five minutes. After cooling to room temperature, add 300 µL of Sodium Potassium Tartrate. The amount of reducing sugars generated was measured using a Jenway 6300 spectrophotometer at 540 nm. The amount of reducing sugars released was determined using the glucose standard curve. One U/mL of FPase represents the amount of cellulase necessary to release 1 µmol of glucose equivalents per minute.
2.8.3. Endoglucanase Assay
Endoglucanase or carboxymethylcellulase (CMCase) activity was determined in triplicates [30]. In 2mL microcentrifuge tubes, 500 µL of 1% CMC was added in 50mM citrate buffer at pH 4.8 [31], followed by 500 µL of crude enzyme filtrate. The reaction mixture was incubated at 50°C for 30 minutes, and the reducing sugars generated were determined using the DNS technique [32]. To halt the reaction, 700 µL of DNS reagent was added. The tubes were then submerged in boiling water for five minutes before adding 300 µL of Sodium Potassium Tartrate. The amount of reducing sugars generated was quantified using a Jenway 6300 spectrophotometer at 540 nm, as described in section 2.8.2. The amount of endoglucanase that released 1 µmol of glucose equivalents per minute is considered one unit of the enzyme.
2.8.4. Exoglucanase Assay
Microcrystalline cellulose (Avicel) (1.25%w/v) was suspended in 100 mM Sodium acetate buffer (pH 4.8), which acted as a substrate. Five hundred (500) µL of Avicel suspension solution was added into 2 mL microcentrifuge tubes, and 500 µL of the enzyme extract was added, and the reaction mixture was then incubated in a water bath at 50°C for two hours. The amount of the reducing sugars produced was resolved as in 2.8.2 using the DNS method [33]. The 1 µmol of glucose equivalents per minute was regarded as the amount produced by one unit of enzyme.
2.8.5. Beta-glucosidase Assay
Beta-glucosidase enzyme activity was determined following the method of Fan et al., [34] in triplicates. One hundred microliters of 5 mM p-nitropheny1-β-D-glucopyranoside (pNPG) prepared in 100 mM sodium acetate buffer, pH 4.8, was mixed with 400 µL of 100 mM sodium acetate buffer, pH 4.8 and 100 µL of the crude enzyme filtrate. The mixture was subjected to incubation at 50°C for 15 minutes. The reaction was terminated by adding 800 µL of 100 mM glycine buffer, pH 10.8 solution, and the release of p-nitrophenyl was quantified at 420 nm using Jenway 6300 spectrophotometer [35]. The resulting p-nitrophenol amount was determined using a standard curve of ρ-nitrophenol. The amount of β-glucosidase giving rise to 1µmol of ρ-nitrophenol per minute was considered to be 1 IU/mL.
2.9. Data Management and Analysis
For enzyme activity absorbance data reading was converted into International Units (IU) using glucose or p-nitrophenol standard curves using the following formulae [18, 27]:
i. International Units (IU) for FPase, Endoglucanase or Exoglucanase
Activity = ((Final abs-c/m) x (df/sample volume) x (1/time) x (1000/180.16))
ii. International Units (IU) for β-glucosidase= ((Final abs-c/m) x (df/sample volume) x (1/time) x (1000/139.1))
Where c= intercept, m= slope obtained from glucose or ρ-nitrophenol standard curves abs = absorbance value df = dilution factor
The data was entered in an Excel spreadsheet and imported into R-software version 3.5.1 (R. Core Team, 2018). The data was first checked for agreement with parametric assumption. Descriptive statistics were then calculated and expressed as mean ± SEM. The effects of substrate, incubation time, and substrate pretreatment on enzyme production were analyzed using one-way ANOVA at p<0.005 significance level. Tukeys Post Hoc Test was used to determine the presence or absence of significant differences among treatment groups. Sequence similarity searches were carried out using the BLAST nucleotide reference database for molecular data analysis. Phylogenetic analysis and multiple sequence alignment using CLUSTAL W were performed using The MEGA software version X program [18, 36] and R version 3.3.2 statistical software. Data was presented using tables and figures.
3. RESULTS
3.1. Isolation and Screening of Fungal Cultures for the Production of Cellulase
Phlebiopsis sp. showed a large zone of clearance on CMC agar (Fig. 1) and was subcultured on PDA medium in mass for use in the study.
3.2. Genomic DNA Isolation, Amplification and Sequencing
The DNA samples were amplified and the PCR results were visualized as shown below (Fig. 2).
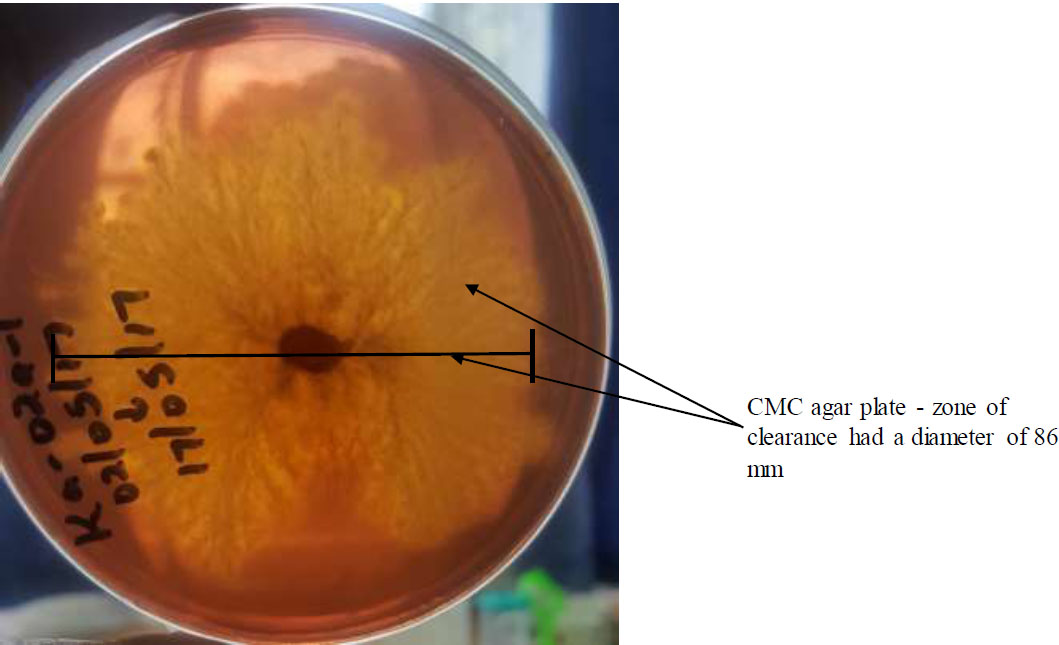
(Phlebiopsis sp.) growing on CMC agar medium.
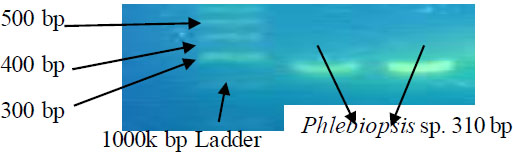
Gel picture of amplified PCR products of Phlebiopsis sp. (310 bp) with 1Kb DNA Ladder in Sybr green-stained gel under UV illumination.
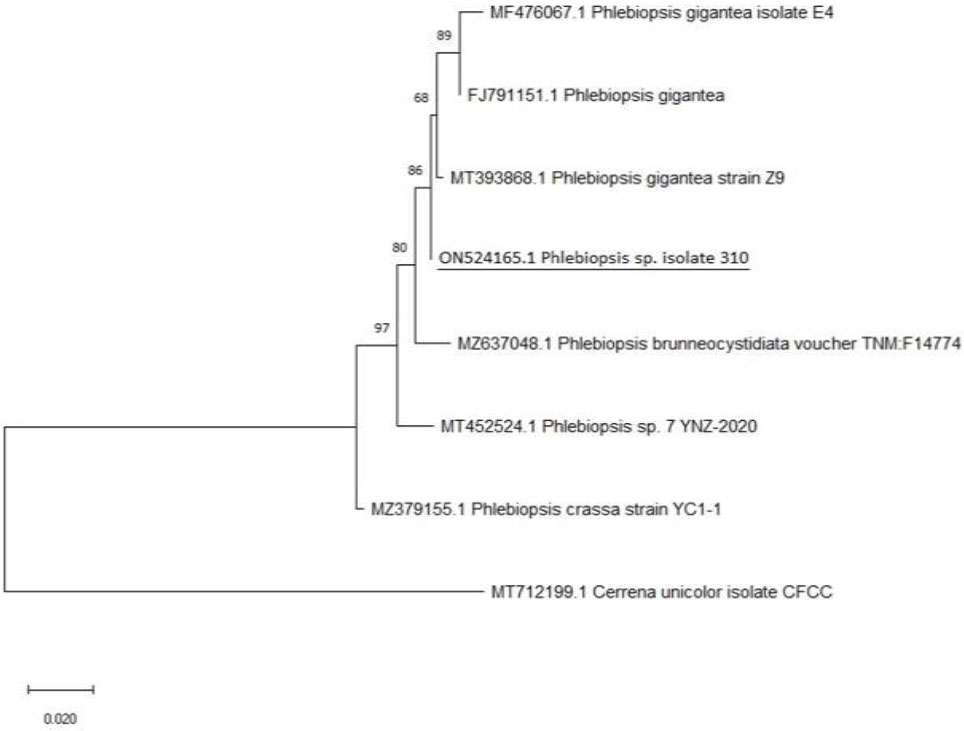
Phylogenetic tree for C1-Phlebiopsis sp. isolate 310.
The isolated fungus was identified as Phlebiopsis sp. (310 bp). Fig. (3) depicts the Neighbor-Joining approach, which was used to determine the evolutionary history of Phlebiopsis sp. The Tamura 3-parameter approach was used to compute evolutionary distances, which are expressed as base substitutions per site. The first, second, third, and noncoding codon positions were all included. The bootstrap consensus tree built from 500 iterations depicts the evolutionary history of the organisms under study. Branches from partitions that have been replicated in fewer than half of the bootstrap replicates are collapsed. Next to the branches are the percentages of duplicate trees in which the related species grouped in the bootstrap test (500 repetitions) [18, 37].
3.3. Effect of Incubation Period on Cellulase Production
The Phlebiopsis sp. produced cellulase differently as a result of the incubation duration.
3.3.1. Effect of Incubation Period on Filter Paper Activity
Table 1 shows that Phlebiopsis sp. produced the maximum FPase production on sugarcane bagasse (35.3 ± 1.28 IU/mL) on day 3, which decreased dramatically on day 6 and subsequently rose on days 9 and 12 to comparable levels on day 3. Maize cobs had the highest FPase activity (33.4 ± 1.85 IU/mL) on the 3rd day of incubation, while rice husks had a significantly higher FPase activity (23.7 ± 2.73 IU/mL) on the 12th day of incubation compared to the 3rd, 6th, and 9th days.
3.3.2. Effect of Incubation Time on Exoglucanase Activity
According to Table 1, Phlebiopsis sp. generated the most enzymes (12.3 ± 1.15 IU/mL) on sugarcane bagasse on the third day, which was substantially greater than on the ninth and twelfth days. The greatest exoglucanase activity in maize cobs was 7.55 ± 0.49 IU/mL on the 12th day, which was not substantially different from the 6th and 9th days of incubation. On rice husks, the greatest exoglucanase activity of 5.96 ± 0.04 IU/mL was recorded on the third day, which was only slightly greater than that reported on the sixth day of incubation.
3.3.3. Effect of Incubation Time on Endoglucanase Activity
Phlebiopsis sp. produced the most cellulase and endoglucanase activity on sugarcane bagasse on day 12 (74.3 ± 0.27 IU/mL) based on the data in Table 1, which was only slightly higher than the 9th day. On day 3, maize cobs had the maximum endoglucanase activity (59.6 ± 1.69 IU/mL), which was substantially greater than on the 6th, 9th, and 12th days of incubation. Rice husks showed considerably increased endoglucanase activity (20.2 ± 4.07 IU/mL) on the 12th day of incubation.
3.3.4. Effect of Incubation Period on β-glucosidase Activity
Table 1 indicates that Phlebiopsis sp. had the maximum β-glucosidase activity (23.7 ± 1.86 IU/mL) on the third day of incubation on sugarcane bagasse, which was only significantly greater than that on the sixth day. The greatest β-glucosidase activity in maize cobs was 22.8 ± 0.18 IU/mL, with no significant difference seen between the 9th and 12th days of incubation. The activity of β-glucosidase on rice husks did not vary substantially between the third and ninth days of incubation nor between the sixth and twelfth days.
3.4. Effect of Substrate Pretreatment on Cellulase Production
The effect of pretreatment showed variation in cellulase production by Phlebiopsis sp.
3.4.1. Effect of Substrate Pretreatment on Filter Paper Activity
The highest filter activity, as shown in Table 2, was recorded on maize cobs pretreated with 0.25 M NaOH of 10.7 ± 0.04 IU/mL for Phlebiopsis sp. Filter paper activities recorded for sugarcane bagasse pretreated with H2O, 0.1 M H2SO4, 0.1 M HCl and 0.25 M NaOH were not significantly different, while for pretreated rice husks, FPase was low and did not differ significantly among the pretreatments.
3.4.2. Effect of Substrate Pretreatment on Exoglucanase Activity
For Phlebiopsis sp., Table 2 shows the highest exoglucanase activity recorded on sugarcane bagasse and maize cobs pretreated with 0.25 M NaOH were 6.14 ± 0.06 IU/mL and 6.17 ± 0.04 IU/mL, respectively. The exoglucanase activities were significantly higher than exoglucanase activities recorded when the same substrates were pretreated with hot H2O, 0.1 M H2SO4, and 0.1 M HCl. Exoglucanase activity recorded on rice husks was relatively low and did not differ significantly among the pretreatments.
3.4.3. Effect of Substrate Pretreatment on Endoglucanase Activity
The highest endoglucanase activity for Phlebiopsis sp. in Table 2 was recorded in maize cobs and sugarcane bagasse pretreated with 0.25 M NaOH at 25.1 ± 0.61 IU/mL and 24.8 ± 0.97 IU/mL respectively. The endoglucanase activities were significantly higher than endoglucanase activities recorded when the same substrates were pretreated with hot H2O, 0.1 M H2SO4, and 0.1 M HCl. Endoglucanase activity recorded on rice husks was relatively low and did not differ significantly among the pretreatments.
3.4.4. Effect of Substrate Pretreatment on β-glucosidase Activity
β-glucosidase activity was exceptionally high among all the pretreated substrates, as shown in Table 2. The highest β-glucosidase activity for Phlebiopsis sp. was recorded on both sugarcane bagasse and maize cobs pretreated with hot H2O at 117 ± 2.52 IU/mL and 118±0.42 IU/mL while pretreated rice husks with 0.1 M H2SO4 recorded the highest β-glucosidase activity of 38.9± 4.95 IU/mL.
4. DISCUSSION
The study involved screening of Phlebiopsis sp. growing on decaying wood from the Kangemi-Loresho area to investigate if they had the potential to produce cellulases. Phlebiopsis sp. was identified using molecular techniques. Phlebiopsis sp. was evaluated on the extent of cellulolytic enzyme production using untreated and pretreated maize cobs, sugarcane bagasse, and rice husks as substrates.
| Cellulase Activity | Substrate | 3 Days | 6 Days | 9 Days | 12 Days |
|---|---|---|---|---|---|
| Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | ||
| Filter Paper Activity | SB | 35.3 ± 1.28aB | 0bA | 30.8 ± 0.79aC | 30.6 ± 3.44aA |
| MC | 33.4 ± 1.85aB | 18.7 ± 2.27aB | 22.0 ± 1.70aB | 28.7 ± 4.00aA | |
| RH | 4.49 ± 1.26aA | 0aA | 12.1 ± 0.54bA | 23.7 ± 2.73bcA | |
| Exoglucanase Activity | SB | 12.3 ± 1.15aB | 10.9 ± 0.23abC | 6.52 ± 0.76bA | 7.41 ± 1.47bA |
| MC | 5.13 ± 0.50aA | 7.11 ± 1.08aB | 6.70 ± 0.37aA | 7.55 ± 0.49aA | |
| RH | 5.96 ± 0.04aA | 0.09 ± 0.07bA | 5.84 ± 0.45aA | 5.59 ± 0.12aA | |
| Endoglucanase Activity | SB | 74.3 ± 0.27aC | 66.3 ± 4.48aC | 43.5 ±5.98bB | 73.9 ± 0.63aC |
| MC | 59.6 ± 1.69aB | 48.6 ± 0.70bB | 43.6 ± 2.20bB | 45.1 ± 1.83bB | |
| RH | 6.72 ± 0.85aA | 17.1 ± 1.64bA | 4.18 ± 0.58aA | 20.2 ± 4.07bA | |
| β-glucosidase Activity | SB | 23.7 ± 1.86aB | 8.82 ± 1.01bA | 18.3 ± 1.31aA | 21.9 ± 1.52aB |
| MC | 22.8 ± 0.18aB | 9.77 ± 0.34bA | 19.4 ± 0.19aA | 18.2 ± 0.46aB | |
| RH | 15.7 ± 0.59aA | 9.63 ± 1.32bA | 16.6 ± 0.26aA | 12.7 ± 1.29abA |
| Cellulase Activity | Substrate | H2O Treatment 121°C | 0.1M H2SO4 Treatment 121°C | 0.1M HCl Treatment 121°C | 0.25M NaOH Treatment 121°C |
|---|---|---|---|---|---|
| 12 Days | 12 Days | 12 Days | 12 Days | ||
| Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | ||
| Filter Paper Activity | SB | 6.33 ± 0.31aC | 7.11 ± 1.84aAB | 7.98 ± 1.33aAB | 7.66 ± 1.79aB |
| MC | 4.74 ± 0.31aB | 5.26 ± 0.45aA | 5.21 ± 0.30aA | 10.7 ± 0.04bB | |
| RH | 2.5 ± 0.07aA | 2.01 ± 0.64aA | 1.81 ± 0.44aA | 2.86 ± 0.33aA | |
| Exoglucanase Activity | SB | 2.76 ± 0.22aC | 0bA | 2.80 ± 0.13aB | 6.14 ± 0.06abB |
| MC | 0.81 ± 0.15aB | 0aA | 1.97 ± 0.34bB | 6.17 ± 0.04bcB | |
| RH | 0aA | 0.03 ± 0.03aA | 0aA | 0.84 ± 0.01bA | |
| Endoglucanase Activity | SB | 11.6 ± 0.31aC | 3.86 ± 0.49bA | 14.0 ± 0.13aC | 24.8 ± 0.97cB |
| MC | 5.62 ± 0.59aB | 4.31 ± 0.19aA | 8.98 ± 0.29bB | 25.1 ± 0.61cB | |
| RH | 2.87 ± 0.63aA | 2.99 ± 0.38aA | 1.58 ± 0.26aA | 1.49 ± 0.18aA | |
| β-glucosidase Activity | SB | 117 ± 2.52aB | 116 ± 0.93aB | 117 ± 1.84aB | 104 ± 2.21bC |
| MC | 118 ± 0.42aB | 114 ± 0.11bB | 118 ± 0.44aB | 95.5 ± 0.72abB | |
| RH | 33.2 ± 1.80aA | 38.9 ± 4.95aA | 38.7 ± 3.04aA | 15.7 ± 2.28bA |
The outcomes reveal that utilizing untreated maize cobs, sugarcane bagasse, and rice husks as substrates, Phlebiopsis sp. produced the entire set of enzymes for the cellulase system. The effect of time of incubation on untreated maize cobs, sugarcane bagasse, and rice husks as substrates for cellulase production by solid-state fermentation was significant for Phlebiopsis sp. Endoglucanase, exoglucanase, and β-glucosidase enzymes are required to work together for cellulase to successfully hydrolyze cellulose [38]. Each of these enzymes cannot single-handedly hydrolyze the complex crystalline cellulose efficiently, but if they work synergistically with each other then they can increase the rate of cellulose degradation significantly [39].
Cheap and readily available plant biomass, such as agro-waste, agro-industrial wastes, various types of grass, cotton stalk, rice husks, maize cobs, nut cakes, sugarcane bagasse, fruit pulp, crop residues, and many others [40], can be used to produce cellulolytic enzymes for industrial applications, including biofuel production. This study found that using untreated sugarcane bagasse as a substrate in solid-state fermentation resulted in the highest cellulase production [41]. Endoglucanase enzyme generated by Phlebiopsis sp. exhibited the maximum activity on sugarcane bagasse, which is comparable to that of T. reesei, which revealed that the optimal incubation period for endoglucanase development on kallar grass [42]. Even when the medium is the same, the optimal incubation period for endoglucanase activity differs depending on the substrate [43]. This might be because varied macro- and micronutrient depletions in the fermentation medium throughout time stressed the fungi's physiology and led the endoglucanase enzyme's secreting mechanism to become inactive [44].
Contrary to the performance of endoglucanase activity, exoglucanase activity was the lowest among all the enzymes, with the best-performing substrate in their production being sugarcane bagasse. These results were similar to Kalsoom’s study on Trichoderma reesei, which showed that the optimum day for exoglucanase production was day 4 on kallar grass [40]. This variation in the incubation period for exoglucanase and low production may be due to cellobiose accumulation at different concentrations, causing feedback inhibition [45].
Sugarcane bagasse was the best substrate for filter paper activity and β-glucosidase activity but was produced in low levels among the substrates as compared to the other enzymes. β-glucosidase activity results are similar to that of a study on Trichoderma reesei by Chandra, who observed that T. reesei has a poor β-glucosidase performance compared to the other cellulolytic enzymes [46]. This is contrary to studies performed on Aspergillus niger whose outcome shows that it has excellent β-glucosidase producing capabilities [47].
This study showed that the endoglucanase, exoglucanase, and β-glucosidase enzymes were produced by Phlebiopsis sp. To create a multi-enzyme system for cellulose hydrolysis, all of the cellulases must interact. Additionally, coordination is needed for the production of the end product and the catalytic movement of the cellulose chain [48]. The high performance of endoglucanase activity may have affected the performance of exoglucanase activity, β-glucosidase and filter paper activity. Both endoglucanase and exoglucanase are major parts of the microbial cellulose degradation system because they both release cellobiose units from cellulose chains [49]. The high activity of endoglucanase may have led to cellobiose accumulation, which caused feedback inhibition thus affecting the activity of exoglucanase, FPase, and β-glucosidase activities [50]. These results indicate that low production of β-glucosidase can restrict cellulase activity while both endoglucanase and exoglucanase enzymes play a vital role in the total cellulase activity [51].
Pretreatment is a key step in the biochemical conversion of lignocellulosic materials into bioethanol because it changes the composition of cellulosic biomass, allowing enzymes that break down cellulose into fermentable sugars [52] to access it more readily [53]. The substrates were treated with hot water, 0.1 M sulphuric acid (H2SO4), 0.25 M sodium hydroxide (NaOH), and 0.1 M hydrochloric acid (HCL) at 121ºC 15psi for 15 minutes. They were then washed with distilled water to achieve a neutral pH. Pretreated maize cobs, sugarcane bagasse, and rice husks had different effects on Phlebiopsis sp. when used as substrates for cellulase enzyme synthesis via solid-state fermentation. Pretreatment with 0.25 M NaOH was the most effective for cellulase formation because it caused the substrates to expand, increasing the specific area while decreasing the polymerization and crystallinity ratios. This separated the structural links between lignin and carbohydrates, resulting in a modification of the lignocellulose structure [54].
From the pretreated data, it is clear that pretreatment only led to increased production of β-glucosidase activity. In addition to this, maize cobs were the best-performing substrates. The pretreatment results are similar to that of A. niger which has been the focus of many studies due to its excellent β-glucosidase producing properties [55]. This could be because the amorphous fraction of cellulose was still present and, therefore, better assimilation of Phlebiopsis sp., unlike the pretreated sugarcane bagasse, which may have lost its amorphous fraction of cellulose. This is what may have also resulted in poor production of endoglucanase, exoglucanase, and filter paper activity [41]. In addition to this, washing of pretreated sugarcane bagasse, maize cobs, and rice husks, residual degradation products like phenolic compounds derived from the hydrolysis of lignin during pretreatment, could have inhibited the fungi growth [41] because of their toxic characteristics to the metabolism of microorganisms [41, 56]. Moreover, the study [57] showed that thermal, acid and alkaline pretreatments released high concentrations of inhibitory-by products [41] like formic acetic acids and phenolic compounds in high concentrations, which inhibit fungi growth during fermentation and therefore reduce cellulase production.
One of the primary limitations of this study was financial constraints, which influenced sample size and access to advanced laboratory techniques. Due to budgetary restrictions, we were unable to include a larger sample population, which may affect the generalizability of the findings. Additionally, resource limitations constrained our ability to conduct more comprehensive analyses using high-cost methodologies. However, we mitigated this challenge by optimizing available resources, prioritizing key experimental procedures, and leveraging collaborations with other institutions. Future studies with increased funding could expand on these findings by incorporating larger datasets and more advanced analytical techniques.
CONCLUSION
This study has shown that Phlebiopsis sp., which was isolated from decaying wood, is a cellulose-degrading fungus that can use sugarcane bagasse, maize cobs, and rice husks as carbon sources for cellulase production. The highest cellulase production results for Phlebiopsis sp. were observed when untreated sugarcane bagasse was used as the carbon source, while maize cobs were the most preferred carbon source for pretreatment. In addition to this, the best-performing pretreatment method is 0.25 M NaOH for cellulase enzyme production in the study, but there is a need to look into other pretreatment methods because the untreated carbon sources produced more cellulase enzymes than the pretreated carbon sources except for β-glucosidase activity.
Future research should focus on optimizing pretreatment conditions to enhance enzyme yields, investigating the economic feasibility of large-scale cellulase production using agricultural waste, and assessing the potential industrial applications of Phlebiopsis sp. in biofuel production and other biotechnological processes.
AUTHORS’ CONTRIBUTIONS
The authors confirm their contribution to the paper as follows: G.O.: Study conception and design; E.M.K.: Methodology; C.I.J., R.O.: Draft manuscript. All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| LCB | = Lignocellulosic Biomass |
| CMC | = Carboxymethyl cellulose |
| PDA | = Preparation of Potato-dextrose Agar |
| DNS | = Dinitrosalicylic acid |
| FPA | = Filter Paper Assay |
| NaOH | = Sodium Hydroxide |
AVAILABILITY OF DATA AND MATERIALS
The data is available from me and will be deposited to Zenodo repository. The data supporting the findings of the article is available in the Zenodo repository at https://doi.org/10.5281/zenodo.15422245 reference number is 15422245.
FUNDING
The study was partially funded by my funds and African Development Bank/Government of Kenya grant Number MOHEST/HEST/31/2014-2015.
ACKNOWLEDGEMENTS
Declared none.


